Genetic Trees explained.
Author:
Thomas Hansen
polterguy@hotmail.com
Date: 28. June 2002
What is a genetic tree?
A question difficult to answer without doing lots of arm-waving and "you know
what I mean?" stuff...
But a Genetic Tree Structure might be defined as an n'node Tree Structure with
Binary Tree capabilities!
When I first thought of genetic trees it was to solve a specific problem and
that problem was to syntax check an Edifact file. An Edifact file is a
relational file (XML is a relational file structure) **WITHOUT!** closing tags.
The fact that they don't have closing tags raises a lot of problems, e.g. how
do I know which level to go to when I am finished with a segment group(*1)?!?
I am NOT going to explain the exact approach I had to take to solve the Edifact
problem, but I AM going to explain how I solved the "linear/relational"
problem...
The way I solved it was by coming up with a tree structure that could be
presented both relational like a tree, and linearly like a list. To do this I
took an n'node tree structure and added up linear information. This could have
been done in several ways, I could have used plain index numbers for instance,
but I wanted to have more power and flexibility then that approach.
The solution I came up with was to assign a genetic code to the
ROOT NODE (Alpha Node) which represented some kind of "minimum value" of
possible combinations of genetic sequences, I choose the ASCII character 'a'.
Then when I added a child to the Alpha Node, I took the 'a' and added up
another 'a' so that the genetic code for the Alpha Node's first child became
'aa', then for every node I added as a child to the Alpha Node I increased the
last character of the DNA sequence by one (Alpha's second child has 'ab',
alpha's third child has 'ac' etc...
The whole point is that a child to any given node inherits it's parent node's
DNA and adds up the character representing it's number in it's siblings flock,
e.g. Alpha's second child's third child will get 'abc', se illustration 1.1.
1.1
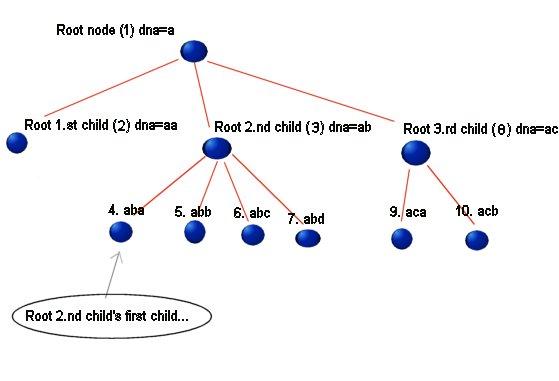
In the figure the first part of the string is the explaining text, the number
in parentheses is the index number of the given node, and the DNA= xx is the
DNA sequence of the given node. (yes I know I forgot the parentheses at the
deepest level of the illustration...Opps! ;) too lazy to fix)
If we explore the possibilities of a Genetic Tree Structure for a
while we will find many really interesting traits that can be exploited by the
implementer:
-
A DNA sequence has got LENGTH according to its depth inside the tree, this
means that we can find out how many ancestors any given node has by examining
the length of a DNA.
-
A DNA sequence LAST CHARACTER will represent a nodes number in it's siblings
flock, e.g. 'afec' as a DNA code mean's that a node is the third children to
it's parent. (the last character 'c' is number three in the alphabet)
-
A node's DNA sequence is actually storing a node's EXACT POSITION into the tree
structure.
-
You can compare ANY GIVEN NODE's DNA sequence to ANY OTHER NODE's DNA sequence
to see "which one comes before the other".
Especially the last trait gives us interesting possibilities, if you can compare
two DNA sequences to see which one is before the other then you can Binary
Partition the tree structure up into a "before this node part" and an "after
this node part"...
You get a fully-fledged Binary Tree Structure without restricting the number of
nodes in each level (plain English:) "You can do a 'binary' search into a tree
where each node may have an 'unlimited' number of children...
The observant reader may be suspicious to the last statement since there are a
limited number of characters from a-z (26 to be exact) which will restrict the
number of siblings a node can have to 25, but really, you don't need to use
character sequences as DNA building blocks, I have merely used them here for
illustration purposes. (it's easier to explain with a string-compare)
If you need a DNA sequence with a higher resolution, you can use e.g. a WORD
structure giving you the possibility to have each node having up to 65 535
children nodes, or even a DOUBLE WORD giving you more then 4 billion
children....
Mark also one very nice feature, you don't have to insert nodes according to
their values like you have to do in a Binary Tree Structure.
Plus the lookup function will go FASTER then in a Binary Tree (as long as you
have the genetic code for what you are looking for) because a Binary Tree will
probably have a much deeper depth then a Genetic Tree and the lookup into which
children node you're going to is just an offset into an array of nodes, off
course you MUST have the genetic code for what you are looking for or the
lookup will have to traverse potentially the WHOLE TREE to find what it's
looking for...
Fact is I believe they in practise for tree structures with very many siblings
may outperform a hash table, but that's NOT the point, the point is that in a
Genetic Tree Struct you have data stored BOTH linearly AND relationally!!!
Ok, this is quite awesome, what's the catch?!?
Yes there is a catch, not one but MANY indeed, although I haven't researched
all of the possible scenarios what's pretty clear to me is that if you generate
the DNA key on the fly while inserting (as opposed to after you have a complete
tree) you will have difficulties to insert nodes in-between other sibling
nodes...
Why is that so?!?
Because you would invalidate ALL DNA sequences to the 'younger' siblings and to
the children to the 'younger' siblings too meaning that you would either get a
corrupt tree structure or you would need to traverse the tree updating all DNA
sequences to the 'younger' siblings and their children etc... (claims a lot of
resources)
e.g. if you insert at 'acb' and you have a node 'acb' from before and an 'acc'
node you would invalidate the DNA sequence to the former 'acb', 'acc', all of
'acb''s children/grandchildren etc... and the same for 'acc'... (MARK! The
sequence up till 'aca' or the sequence from 'ab' and out would NOT be affected)
However there are several workarounds this problem, you can either restrict the
insertion points to consequently being a 'back_insert_child()' /
'insert_youngest_sibling()' which would completely eliminate the problem by
narrowing the possibilities, or you could generate the DNA sequence for the
tree AFTER you are done inserting nodes through some kind of
'validate_DNA_for_tree()' function.
There are other bad scenarios too, but most of them have a foundation in the
fact of that the insertion point is either restrictive or potentially
expensive...
MARK!
Only the 'Family Tree' of the 'younger siblings' of the insertion point will be
affected by an insert...
What are they good for?!?
As I said earlier, if you need to be able to have data (works best with
'static' or 'read once' data) in BOTH a relational and a linear representation,
Genetic Trees are the way to go.
E.g. take an XML Schema file for instance, you read it once in some kind of
'read_schema()' function and you need to know which node is a children to which
other node (relational part), plus you need to know which node comes before
another node (linear part) because you must have XML tags in a certain order
for the XML document to be valid against the Schema.
But I have only used them in the Edifact domain, so I wouldn't have a clue
about the applications they could be of use in other then the ones mentioned.
But if you need to represent data in both a linear and a relational way without
managing duplicated data structures, Genetic Trees are the way to go.
(*1) A segment group in an Edifact file is "ONE LEVEL" of segments
where a segment is "one tag" containing data specific to that tag, e.g. the
"BODY" tag of an HTML document is one level in an HTML document even though the
BODY tag can contain inner levels e.g. a "P" element (P=paragraph) or an "I"
element (I=Italics)
GenTree.h
I have in fact implemented a Template Container Class for manipulating Genetic
Tree Structures, I haven't been working too much on the classes, roughly about
24 hours of programming, but I think they are versatile and stable enough to
get the idea, and maybe to do a sample implementation of an "n'node tree
structure with Binary Tree capabilities".
They are implemented in C++, built and tested with Microsoft Visual Studio 7.0.
(I really missed partial template specialization)
But I think they will compile in most of todays compilers.
They use the "iterator design pattern" (STL way), and allthough I have tried it
with a couple of the std algorithms, I guess they will tremble upon some of
them, especially since the end iterator is weakly implemented, good ideas or
refactoring of the end iterator is greatly appreciated!! (it's a special value
today)
All though they are implemented in C++, I think most OOP capable programmers
with some "iterator knowledge" and basic knowledge about generic containers
will be able to port it to their favourite language...
Download sample Generic Genetic Tree Template library for
C++ (gen_tree.h) and sample use of gen_tree.h (gen_tree_tester.cpp) contained
in 'gen_tree.zip'
If you would like to comment about Genetic Tree Structures, my implementation
of them(gen_tree.h), my bad English or anything else presented in this article,
please don't hesitate to mail me at polterguy@hotmail.com
BTW!
If you're looking for a very good template based C++ Windows API GUI library have a look at my main project SmartWin
And if you're interested in other things I'm thinking about at the moment, check out my blog HERE or my current GUI library here... ;)
Thomas Hansen
"By the time we've finished with him, he won't know whether he's Number Six or
the cube root of infinity!"
