Finding the Hypermutation Consensus Sequence:
The Next Step
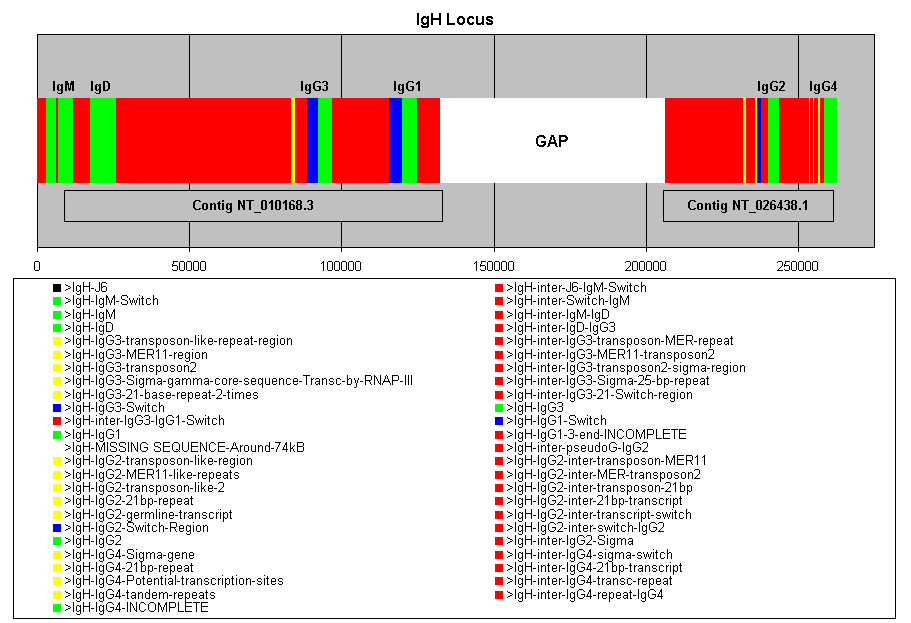
A brief description of the annotated loci
The publicly available sequence has been obtained
and annotated. In other words, the entire Ig kappa and Ig lambda loci are
now annotated, as is approximately half of the Ig heavy locus.
The heavy locus was found on two separate contigs.
There was a gap between the two contigs, and it was calculated to be approximately
75000 base pairs. The 3' end of the Ig heavy locus is also missing. Thus,
only the IgM, IgD, IgG3, IgG1, IgG2 and IgG4 regions are available. The
gap encompases the pseudoE, IgA1 and pseugoG genes, whereas the IgG and
IgA2 regions are in the missing 3' end.
In the heavy locus, I was able to obtain the following
complete intergenic regions: IgM-IgD, IgD-IgG3, IgG3-IgG1 and IgG2-IgG4.
There are two incomplete intergenic regions: 8 kB in the 5' end out of
19 kB of the IgG1-IgPseudoE region, and 25 kB in the 3' end out of a possible
40 kB of the IgPseudoG-IgG2 region. As one of the incomplete regions
contains only the 5' end, while the other has the 3' end, it is likely
that at least one of these regions will not contain the HCS, if we assume
that the HCS has a conserved position in the intergenic sequence. Or for
that matter, if we assume that the HCS is actually present in the intergenic
region. Or for that matter we have to assume tthat the HCS exists at all...
That's a lot of assumptions indeed...
Summary of the loci, and the proposed experiments
In this section, I will only deal with the experiments
concerning the region 3' of the IgC, and not the intronic regions. This
is not to say that the intronic regions will not be analyzed at some point,
but at this point I feel that there is a higher probability that the HCS
is in the 3' region (see Scharff and Neuberger articles).
The hypothesis of this project is that the HCS is
present in the region 3' of the constant gene. In this locus, there are
4 such complete sequences, and 2 incomplete sequences. However, IgM and
IgD are translated from the same transcript, and it is thus likely that
there is one HCS that controls the hypermutation of both transcripts. Hever,
it's unknown whether the HCS is in the IgM-IgD region, or 3' of IgD. Therefore,
3 parallel experiments need to be performed: one comparing both sequences
to the rest, and the other two comparing only one of the sequences to the
rest (Expts 1-3; see table below for summary of experiments).
Another question that arises is whether the incomplete
sequences (3' of IgG1 and 5' of IgG2) should be used in the comparison.
As the regions are incomplete, the HCS might not be present, even if the
hypothesis is correct. This introduces even more uncertainty, and these
regions ill not be used in the preliminary experiments (Expts 4-7).

The IgK locus is complete. The 3' region contains
a potential transcription unit similar to the BENE gene. The NKG2E gene
is even further 3' downstream. This poses a question of whether the last
part of the 3' region (between BENE and NKG2E) should be used in the analysis.
The IgKC-BENE region encompases around 40 Kb. While it's possible
that the HCS is even further downstream of that, it's unlikely, and therefore
only the 40 Kb will be anayzed initially.
In Entrez, the size of the MAR was said to be 1000
bp. This seems more like an estimate, rather than an actual analysis, and
it should be verified at some point in time...
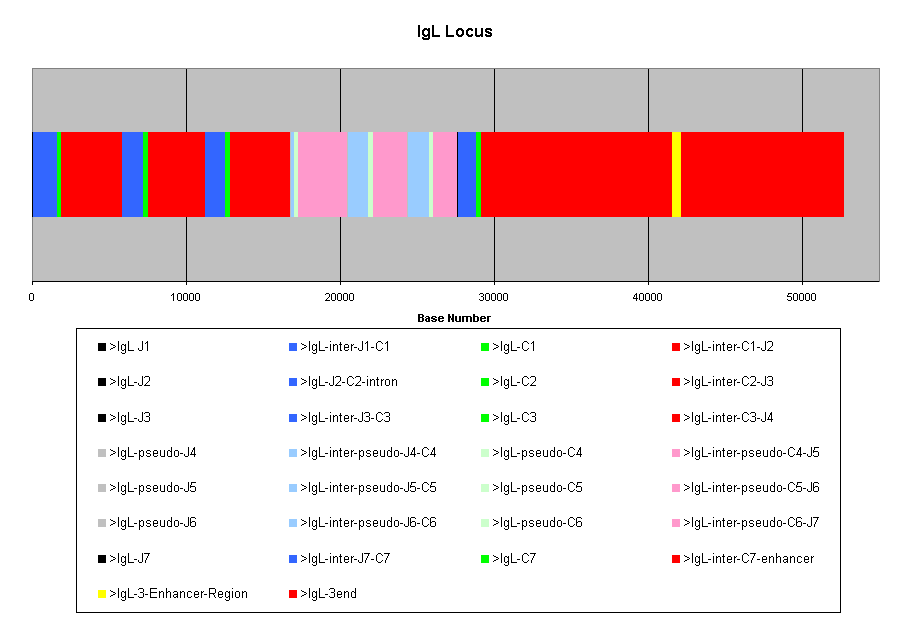
There are 4 IgL C genes and 3 pseudogenes in this
locus. The "real" genes have relatively conserved intronic sequences, different
from the pseudogenes. This suggests a duplication event in the locus. The
region analyzed will consist of the 3' end of the locus, from C7 to the
end, and including the enhancer. There is a question of whether the enhancer
should be included, since it may contain repetitive sequence, or other
regulatory sequences that might be found in the oher loci. However, it
will be included for now, and if it turns out that there's too much info,
I will exclude it.
Table of intended experiments
|
EXPT #
|
IgM-IgD
|
IgD-IgG3
|
IgG3-IgG1
|
3' IgG1
|
5' IgG2
|
IgG2-IgG4
|
IgLC7-enh
|
IgL ENH
|
3'IgL ENH
|
IgK-BENE
|
3' BENE
|
|
1
|
X
|
X
|
|
|
|
|
|
|
|
|
|
|
2
|
X
|
-
|
|
|
|
|
|
|
|
|
|
|
3
|
-
|
X
|
|
|
|
|
|
|
|
|
|
|
4 (NO)
|
|
|
X
|
X
|
X
|
X
|
|
|
|
|
|
|
5 (NO)
|
|
|
X
|
X
|
-
|
X
|
|
|
|
|
|
|
6 (NO)
|
|
|
X
|
-
|
X
|
X
|
|
|
|
|
|
|
7
|
|
|
X
|
-
|
-
|
X
|
|
|
|
|
|
|
8 (NO)
|
|
|
|
|
|
|
|
|
|
X
|
X
|
|
9
|
|
|
|
|
|
|
|
|
|
X
|
-
|
|
10 (NO)
|
|
|
|
|
|
|
|
|
|
-
|
X
|
|
11
|
|
|
|
|
|
|
X
|
X
|
X
|
|
|
|
12 (NO)
|
|
|
|
|
|
|
X
|
-
|
-
|
|
|
|
13 (NO)
|
|
|
|
|
|
|
X
|
-
|
X
|
|
|
Thus there are 4 groups of variable experiments, and
each experiment in the group should be performed with all the possible
alternatives. This gives a total numbr of experiments of : (3) x (4) x
(3) x (3) = 108! This is a bit much, and some of the scenarios are more
likely that others.
For example, experiments 8 and 10 are largely unnecessary,
as BENE transcript likely marks the end of the 3' region of IgK. However,
BENE is a hypothetical transcipt, and perhaps it is inside the 3' IgK region.
This, however, is unlikely. Therefore, experiments 8 and 10 will not be
performed.
Another point of question is the validity of including
the incomplete regions (3' of IgGG1 and 5' of IgG2). As the regions are
incomplete, the HCS might not be present, even if the hypothesis is correct.
Thus, experiments 4, 5 and 6 will also be put on the proverbial shelf for
now.
With regard to IgL, the enhances might contain some
repetitive sequence, or conseved regulatory elements, and thus, it could
be left out. However, the strcture of the IgL locus is unorthodox, and
thus, I feel uncomfortable making too many assumptions with very little
ground to stand on. Thus, I will include the whole 3' region of IgL, and
expts. 12 and 13 will not be performed.
This reduces the number of experiments to 3,
as the only variables are the IgM and IgD regions. To me, it is most likely
that the HCS will not be present in the IgM-IgD region, as IgM and IgD
are a part of the same primary transcript, and there is no switch region
between them (and thus IgM cannot be excised from the genome without excising
IgD - it thus follows that the two are under the control of the same HCS).
Further, the IgM-IgD region is more like an intron, and I'll go out on
a limb and say that the HCS is likely 3' of IgD. Nonetheless, this logic
is shady at best, and thus I will perform the three alternate experiments.
Now, to prepare the sequence...


