Comparison of Whit Athey’s haplotype predictor with
phase 3 37 STR cluster analysis
John McEwan and Whit Athey
13th September 2005
Whit Athey’s haplotype
predictor https://home.comcast.net/~whitathey/predictorinstr.htm
is widely used to predict the likely haplogroup of
STR haplotypes. The methodology used is described in Athey (2005). This methodology has
been extensively tested, and is largely unrelated to the clustering approach
used in the phase 3 analysis. It therefore provides an independent test of the
clustering methodology. A third method is user defined haplogroup
classifications, but while some of these have been validated by SNP testing
many self defined haplogroups are also present, including
several errors. However, they are valuable where conflicts occur between the
methods as the haplotype predictor does not cover all
haplogroups. The potential exists by comparison of
all methods to identify errors either in user haplotype
entry or in user haplogroup labeling.
Method
The approach used was to graph the % haplotype
match estimates from the predictor for each of the defined haplogroups
against cluster order number and individual identifiers listed. Please note
that cluster order has been used here, but it merely indirectly reflects
cluster distance from neighboring branches. The whole cluster was not graphed,
most of R1b was excluded. Haplotype predictor
estimates were calculated in batch mode and the full 37 STRs
were used.
Results
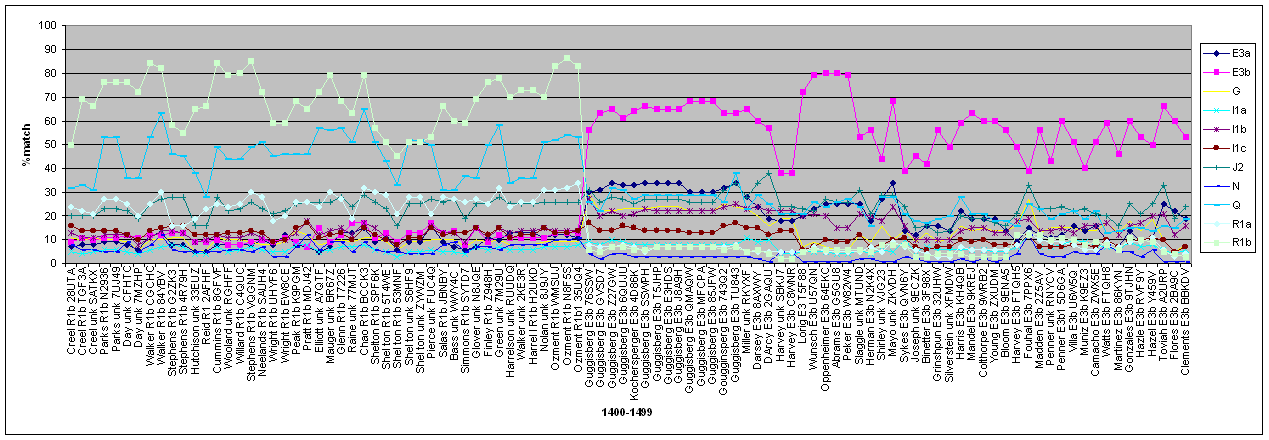
The series of graphs are shown below, each incrementing by 100 haplotypes. The graphs commence at the edge of the initial
R1b cluster and abruptly shift to E3b as is predicted by user defined haplogroups. Subsequently there is an abrupt shift to E3a,
albeit there are several intermediate E3a and E3b predictions in the middle of
the E3b group. Percentage match predictions then drop rapidly for about 9
individuals. These haplotypes where annotated by
users are labeled A, B and C: groups the haplotype
predictor presently does not cover. The J2 haplogroup
then immediately commences and there are two strange haplotypes
which the predictor calls as E3b in the middle of this group. There is then
another abrupt transition to I1c haplogroup albeit
within this group there are periodic haplotypes that
have been clustered within the I1c group, but the predictor cannot make a
reasonable prediction to any group. Unfortunately, none of these individuals
has a user supplied haplogroup prediction. The percentage match prediction then slowly
declines for I1c while I1b begins to rise with a clear cut, but modest
transition. It is noted that one of the user defined haplotypes
in the later part of the I1c regions is defined as I2 and the switch to I1b is
on the observed boundary defined by the predictor. One individual in this group
has a very low % match. There is an abrupt and marked transition to I1a haplogroup. Interestingly, the % match through this entire
region has a high probability for J2, suggesting that the predictor has some
difficulty separating these two groups. At the end of the I1a group is a region
of low % matches and external evidence suggests that this is the group labeled
by others as Ix. An abrupt transition to the G haplogroup
then occurs. Around order number 2570 another poor prediction region commences with several individuals labeled G by the
user predicted as J2. A small bunch of haplogroup F
individuals then appear but are predicted as G with low confidence. It is
interesting these individuals are also clustered together. G % matches then
abruptly decline and until haplotype N % matches
dramatically appear, a confused predictor region exists. According to the user
defined haplogroups this region consists of O3, I, R,
F and K2 haplogroups. Perhaps the most surprising is
the I group and these individuals need further examination as to why they are
clustered away from the I haplogroup and are only
poorly estimated by the predictor. The N
haplogroup is followed by the dramatic appearance of
the Q haplogroup. Strangely, two user and haplotype predictor defined E3b individuals appear on the
interface between Q and R1a. The R1a group is cleanly segregated in their
predictions with the exception of one individual with an extremely low value.
The R1b group then emerges, albeit a small group of Q haplotypes,
including one user annotated haplotype, appear to
space the R1a and R1b groups. It is interesting to note that the R1b haplotype predictions have relatively poor discrimination
between Q and R1b.
In summary, the region examined consisted of more than 1600 haplotypes through the area of greatest diversity, and
these issues were noticed:
·
Small groups were not called by the predictor, as the
relevant haplogroups were not included, but they
still appear to be well clustered by the routine
·
Several individuals that were clustered within a known
haplogroup,
appeared to not be called by the predictor. These have a high chance of being
entered incorrectly, or contain an allele outside the known bounds of the haplogroup in the haplotype
predictor.
·
There are two E3b predicted individuals clustered in
the E3a group, another two in the J2 group and several other cases of clusters
of several individuals from a parent haplogroup
residing in the interface of two larger sub-groups. In some cases it is not
clear what is causing these results, in others it may be as yet undefined
groups or problems due to the clustering method.
·
Ignoring haplogroups that
are not defined by the predictor, and comparing the clustering method and the haplotype predictor identifies that the concordance of the
two methods is extremely high.
·
The overall conclusion is that at the level of well
defined haplogroups the clustering method is probably
more than 99% accurate, throughout the diverse region examined.